It is the time of the year, where our group opens a door at our advents calendar again. It is my turn to open the first door. Our group is involved in many research projects involving genomic data as you can see when looking at our previous posts. In parallel, there are many large-scale genome projects going on like BGE, EBP-Nor, ATLAsea and DToL. Hence, the genomic information, especially of high quality or even reference chromosome level, available for research projects is growing in leaps and bounds. At the beginning many projects concentrated on iconic and model species to make it attractive for funders to invest now. There is a growing understanding by founders and general public that we cannot understand biodiversity and the underlying genomic basis with concentrating on these species only. Accordingly, more and more funding is obtained to address biodiversity at large for the generation of genomic data, for example, at a national level or for certain habitats. The question is now are we getting closer towards the goal or are still biases which biodiversity we cover in these projects.
When one looks at the output of all these efforts there is still a huge bias to certain parts of biodiversity. Despite all the technological progress in the last decade, even the most modern approaches have strong limitations towards the quality and quantity of the DNA. This is often referred to with terms like feasibility. For example, in the BGE project we had a community-based approach. In this approach, the European scientific community could suggest species, for which they could provide samples for the generation of reference genomes. About half of the suggested species were considered not feasible given the feasibility criteria set by the participating sequencing centers. When we look at the figure below, we can see that for some of the phyla from which species had been suggested all were feasible. However, for a similar number of phyla none of the suggested species were feasible. Moreover, as we very openly communicated the feasibility criteria and that these species would not be selected, it can be safely assumed that the numbers of non-feasible species would be higher. This might also include phyla, where in this example all species were feasible. On such example is flatworms, where none of the meiofaunal representatives had been suggested, which would have been considered non-feasible given their small body size. Some phyla consisting of only small-sized species like Gastrotricha were not even suggested in the first place. Hence, what is not feasible and what can we do about it?
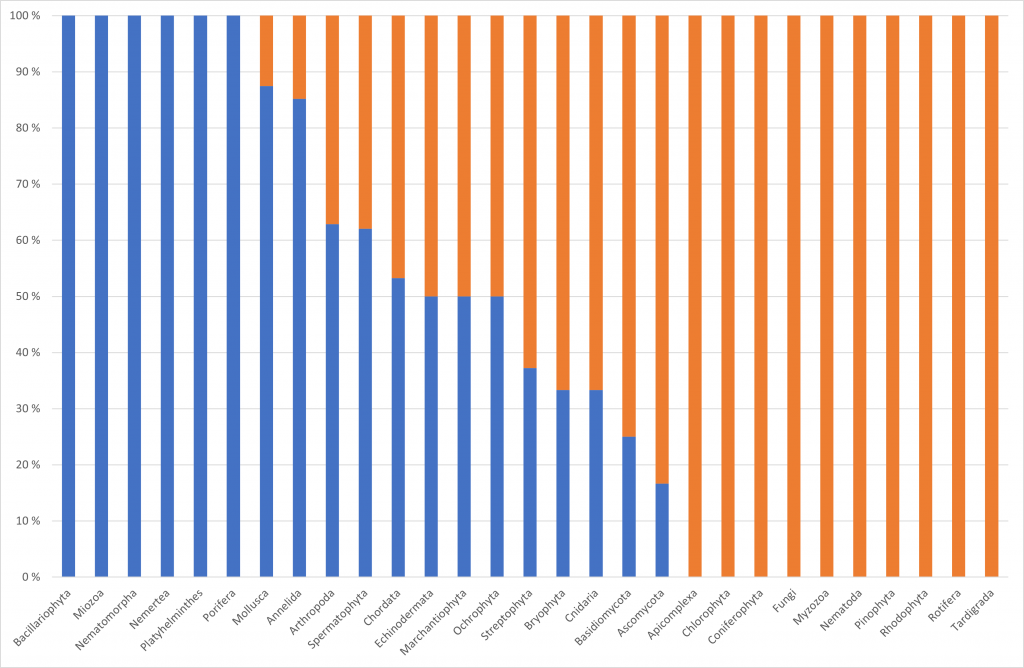
Feasibility criteria checked for different aspects of the selected species and the sampling process to ensure a smoother generation of reference genomes using the available technologies. Given the limited available funding, genome size was one of them as large genome require a large amount of high quality DNA but also a lot of sequencing. Hence, it becomes expensive very quickly. Another criteria is the amount of tissue that can be provided for for the generation of reference genomes in a reasonable time frame. There are meanwhile protocols available allowing to work with much less DNA then a couple of years ago. Nonetheless, all species smaller than for example a fruit fly are still being considered not feasible. Finally, the preservation, storage and transport of the tissue until the extraction of the DNA has to happen at the best possible way ensuring the highest quality of the DNA. This means usually that the tissue is flash-frozen in liquid nitrogen or on dry ice and that afterwards the sample is always kept at -70°C.
Looking at the non-feasible species of the BGE project, we can see that about one fifth had too large genomes, another fifth could not provide material in a reasonable time, about one third a too small sample size and half could not preserve the tissue in an appropriate manner or keep the cold chain. Some species fulfilled more than one of these criteria. This indicates that despite all the technological advancement in sequencing at present, a large proportion of the eukaryotic biodiversity still provides substantial challenges for the generation of reference genomes because they are too small, too rare or the genome too big. Accordingly, more research effort is needed to make the sequencing of reference genomes feasible for a much larger part of the biodiversity.
On the other hand, about half of the species were excluded not due to challenges in the sequencing technology but due to challenges in the sampling procedure. This means that solutions need to be developed that build up the capacity and logistics to preserve and store samples properly for genomic research. For example, access to dry ice or liquid nitrogen need to be accessible allowing timely preservation and short-term storage even in remote areas. Infrastructure for long-term storage needs also to be easily accessible for maintenance of a proper cold chain. Building up these infrastructures and protocols will also be beneficial for species, that are not easy to collect. With such means in place, the threshold to collect species for biodiversity genomic research for non-experts in genomic research would become substantially lower and hence, for example, taxonomists might be more easily convinced to collect specimens for genomics research during their field work just on the possible chance that it might be suitable for sequencing in the future. However, even with all of these in place, for several species it will remain challenging to obtain optimally preserved material due to the fact that they need to be sampled in areas such as the deep sea, remote islands or high-alpine mountainous regions, which are not easily accessible. Hence, development of sequencing protocols for sub-optimal preserved and maintained material will be highly beneficial for such samples.
Taking these considerations into account, if we truly want to sequence a reference genome for each eukaryotic species we cannot proceed as we do right now. From selecting only iconic species we moved to selecting feasible too easily fulfill the expectations by the funders. However, this will result in a strong bias and the research and development of new methods for this biodiversity has to happen now and the same goes for building up the required infrastructure. If we wait with this until we reach the large production phase of genomes we will very quickly exhaust the feasible species and suddenly experience a huge drop in genome production when it should pump out genomes on a daily basis. If we start the development of methods then, it could jeopardize the whole endeavor as the funding agencies would wonder why we cannot deliver to expected output. Moreover, it would provide us with a biased view on the genomic diversity that is our there in the world.
However, there is hope. A few projects like our own InvertOmics project started to delve into some of these challenges. For example, we deliberately choose to work with species of small body size to find ways to get genomes for them. Even we cannot get them to reference chromosome level, we might be able to generate so much information that we get a genome of very high quality at least. Our first successful approach has recently been accepted for publication and will present it in a separate post soon. Additionally, two Master theses are finishing also investigating such species. We will present here the challenges these species provided and the successes the students had after they have successfully there theses. However, these are only small efforts and the community at large needs to put more effort into this. Especially, if one considers that there even more challenges in store also from the species considered feasible. This is also lesson we learn right now from the BGE or other efforts like our InvertOmic project. Nature often throws us a curve ball with new aspects of life.
Hence, biodiversity genomic research is at a crossroads right now. Will we continue to follow only the easy path neglecting substantial parts of biodiversity with all the new knowledge we can gain from it or will we go the bumpy road of endeavoring into uncharted waters with setbacks and strong engagement, both with respect to money and manpower?
Therefore, quo vadis biodiversity genomic research?
![]()

2 Comments on “Door 1: Quo vadis biodiversity genomic research?”