We are happy to announce the first genome release note of the InvertOmics project in Genome Biology and Evolution. The genome of Emplectonema gracile has been sequenced and generated at the reference level of the EBP standards. What does this mean? The quality of the genome is of such high quality that it can be used as reference for other studies using genomic data of less quality and guide the analyses of the data. In this case, it means that we were apply to retrieve each individual chromosome of the specimen we sequenced. Moreover, there hardly any gaps in the chromosomes. Something, which has only been achieved for the human genome not so long ago.

The specimens we took for this study were among the first we collected for the project still during Covid times (see this post). Emplectonema gracile is also called the green ribbon worm as it is a worm of the group Nemertea and it is very green. Ribbon worms produce a lot of slim and this can sometimes be a challenge for genomic studies as this slim might interfere with the experiments in the lab (if you want to learn more about nemertean see this blog post). However, after several trials, Alberto could optimize the protocol to get high quality DNA out the worms. With an combined international effort including people at the museum, at the Norwegian Sequencing Center and the University of Alabama we are able to get the necessary data and the bioinformatic analyses done. This genome was also part of the ERGA pilot genome effort (see blog post)
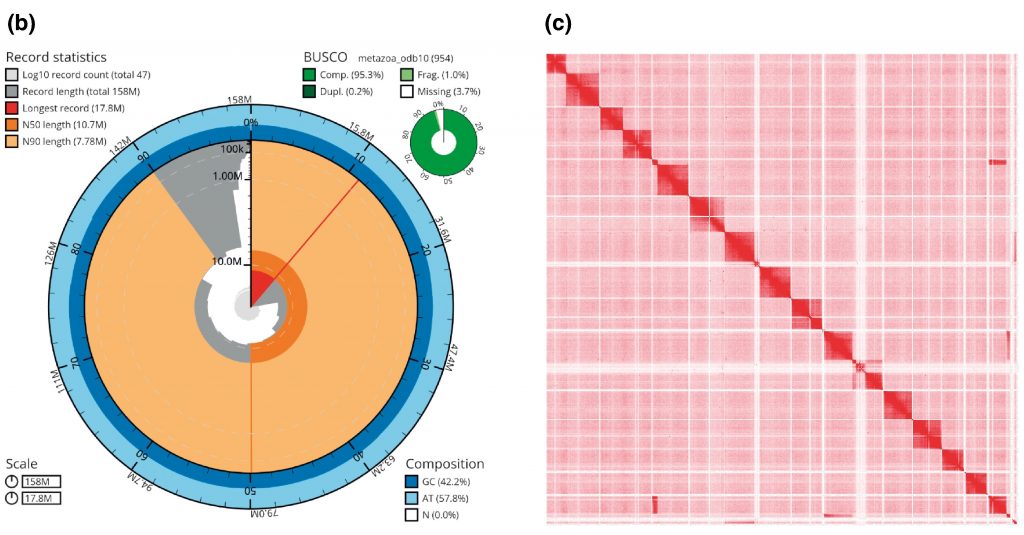
So how does genome look? The genome has a size of 157.9 Mb, which is about one twentieth of the human genome size. It has 17 chromosomal pairs and we could find 95.1% of complete BUSCO genes. BUSCO genes is a set of genes, one excepts to find in animals, and hence it can show who much of the genome probably has been found. Values above 90% are considered the benchmark for a complete genome as not surprisingly one never finds all genes. The annotation of the genome predicted that 20,684 protein-coding genes are in the genome. That is in the same ballpark as for humans.
Alberto Valero-Gracia, Nickellaus G Roberts, Meghan Yap-Chiongco, Ana Teresa Capucho, Kevin M Kocot, Michael Matschiner, Torsten H Struck, First Chromosome-Level Genome Assembly of a Ribbon Worm from the Hoplonemertea Clade, Emplectonema gracile, and Its Structural Annotation, Genome Biology and Evolution, Volume 16, Issue 7, July 2024, evae127, https://doi.org/10.1093/gbe/evae127
![]()
1 Comment on “Slimy but shiny – the first reference-level genome from the InvertOmics project”