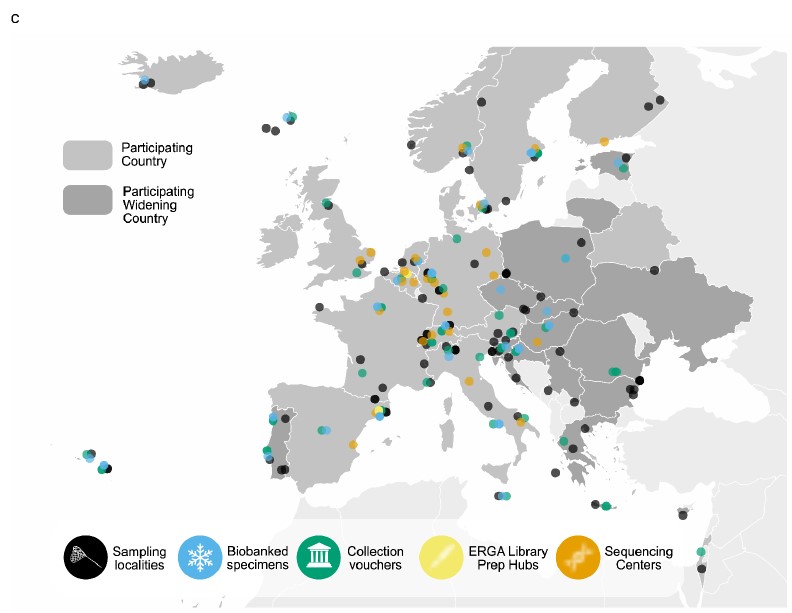
As a proof of principle the European Reference Genome Atlas (ERGA) consortium initiated at the beginning a pilot study. Several sequences centers and different research projects contributed to this pilot study allowing the first sequencing of reference genomes across Europe and setting the stage for application of the Biodiversity Genomic Europe (BGE) project. With our InvertOmics project, we also contributed to this pilot study by sequencing the reference genome of the nemertean Emplectonema gracilis, a green slime worm. While we are still working on the release of this reference genome, we as contributors to the pilot project analyzed the results of the whole process and progress of the pilot so far and what lessons we could learn from it. The paper is now released as a preprint. This great effort both the pilot and the flagship paper were lead by Ann M Mc Cartney, Giulio Formenti, and Alice Mouton. First of all thanks to the great work they did throughout.
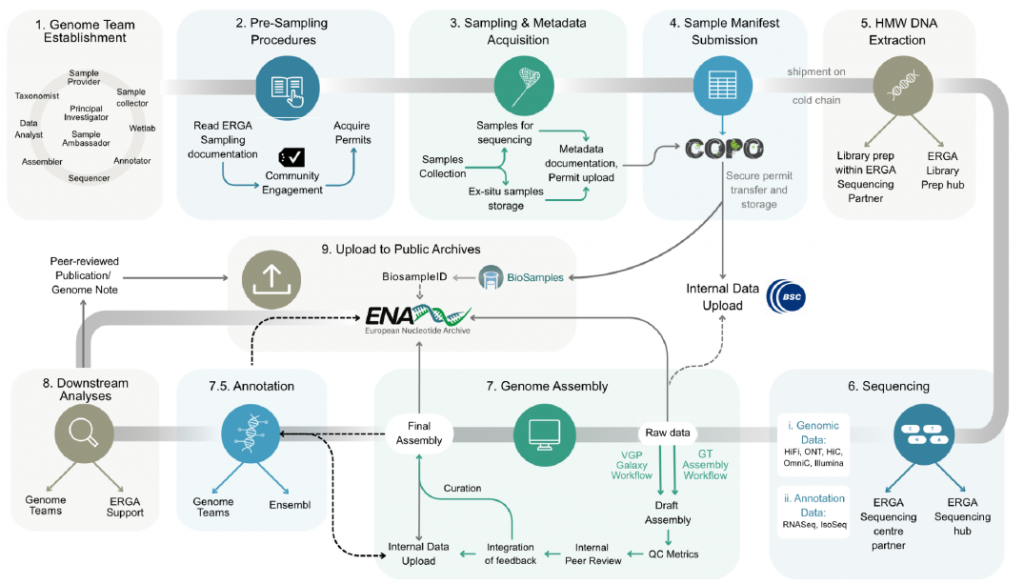
infrastructure that could support the streamlined and scalable production of genomic resources
for all European species.
The paper outlines the process and challenges faced during the development of a pilot infrastructure for the production of reference genome resources, and explore the effectiveness of this approach in terms of high-quality reference genome production, considering also equity and inclusion. The outcomes and lessons learned during this pilot provide a solid foundation for ERGA while offering key learnings to other transnational and national genomic resource projects. The first crucial step was the development of a workflow of decentralized infrastructure from the species selection to the release of the genome bringing together experts from different fields such as taxonomy, molecular biology or bioinformatics. The workflow comprises a total of nine steps and the first four ones relate to the selection process to the sample submission, which we addressed in a separate paper (preprint and blog).
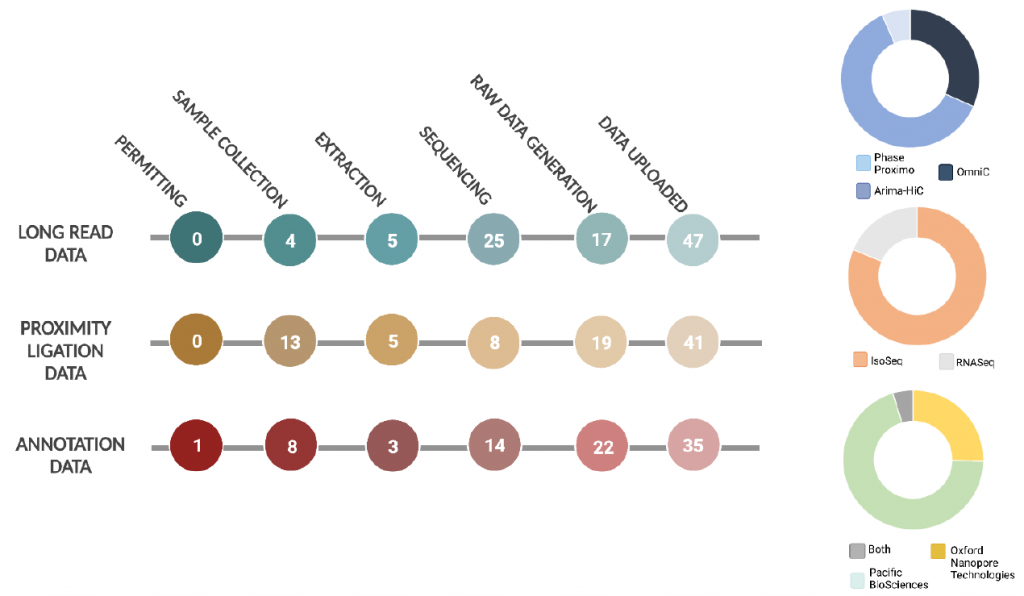
Despite all the progress seen in the figure above the pilot also revealed seven major challenges to a decentralized genomic project like ERGA is.
1) Phylogenetic representativeness and sampling bias
2) Effective engagement due to resource and time limitations, and lack of training and awareness
3) Decentralizing reference production and reproducibility
4) Genome annotation due to lack of sufficient evidence (transcriptomic and protein sequence data) from certain taxonomic groups, databases and predictive models of repeats
5) Ethical and legal compliance, especially across multiple countries even in a setting like the EU provides
6) Training and knowledge transfer with limited resources
7) Building a more inclusive, diverse and equitable infrastructure
The decentralized approach taken by ERGA illustrates the huge potential to become a model for equitable and inclusive biodiversity genomics. The power of such an approach was evident by successfully uniting an international community of biodiversity researchers, but it also stimulated communities of researchers within the same country to combine and consolidate efforts. As ERGA progresses, now with a dedicated funding stream through BGE, it can now build upon, learn and make the intentional investments needed to address at least some of these challenges. Although a centralized source of funding to support these endeavors is overall a positive it will also provide many challenges concerning diversity and equity, however efforts are underway to safeguard at least some level of the decentralized process e.g., community sampling and hot-spot sequencing.
![]()