Last week on April 11th and 12th, the second Norwegian Biodiviersity and genomics conference organized by EBP-Nor took place at Forskningskparken in Oslo. The conference had interesting invited key lectures by Aoife McLysaght, Alexander Suh, Lene Lange and Olga Vinnere Petterson but also our group had a very strong representation at the conference. With 5 talks we contributed almost 22% of all contributed talks and showed the full breadth and depth of research by CEG in biodiversity genomics. From the Natural History Museum only one other contribution was present at the conference, which was not from CEG.
Our contributions kicked off on the first day by a presentation by our Master student Pia Merete Eriksen with the title “Another step up the phylogenetic hill: Assessing the viability of macrosynteny as a phylogenetic trait in Lophotrochozoa”. In her talk, she presented the general topic of macrosynteny used a phylogenetic marker and how she intends to use in her Master thesis to address the still completely unresolved phylogeny of Lophotrochozoa, one of the three large animal groups. This work is also contributing to the goals of the InvertOmics project.
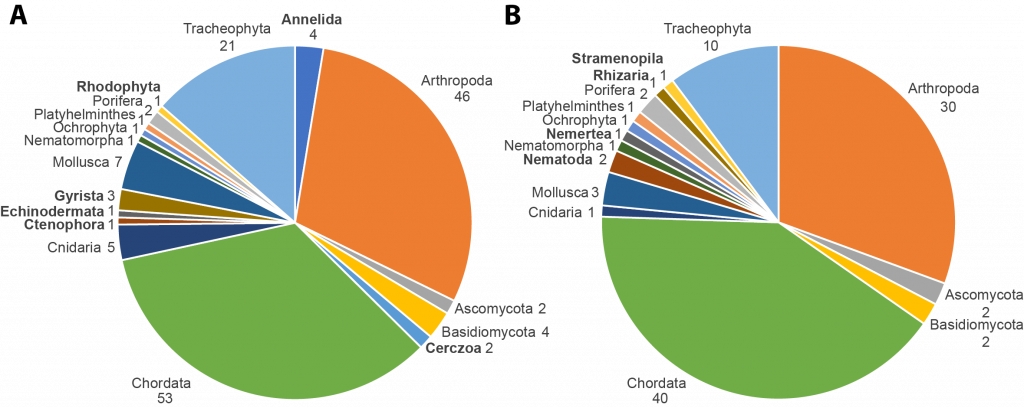
But the second day was truly our day. It started off with a presentation by Torsten Struck (“Species selection for large scale genome projects an automated process based on explicitly bottom up defined criteria”) in which he explained the procedures on how to objectively select species from community suggestions in large-scale international projects such as BGE of CEG is a part off. This also showed the advantage on developing such procedures in a community-driven as it was developed by the ERGA community, which includes several members of the CEG group.

It then continued with the passionate talk by Marvin Choquet (“Unlocking the first large genome of the key zooplankton genus Calanus: challenges and insight”) about doing genomic research in the ecological and also economically important copepod species of Calanus. They present some of the typical obstacles we are facing now with expanding genomic research beyond the usual suspects such as vertebrates, insects and plants. They are very small but have genomes at least twice the times of a humand genome. This research is related to the STADIS PostDoctoral fellowship Marvin is holding.
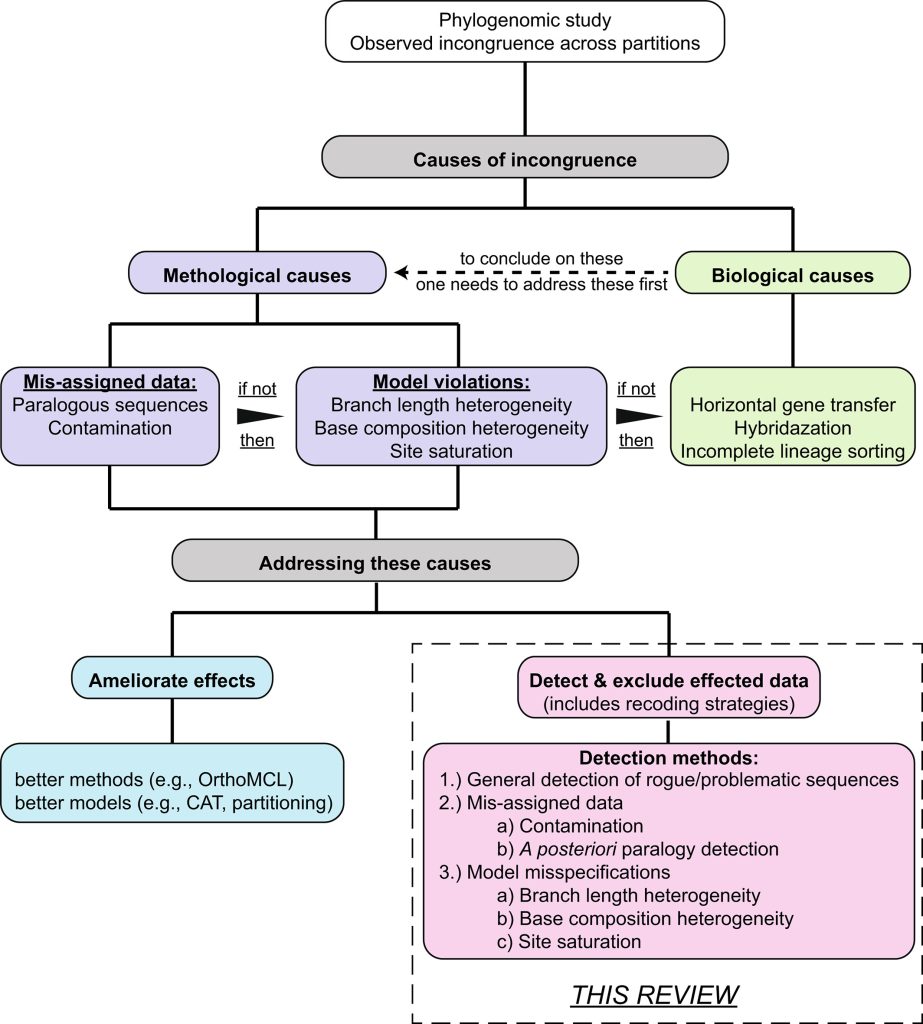
The last two talks then returned back to research done in our InvertOmics project. The first talk was by our PostDoc James Fleming on “BOSTIn – User friendly software to assess potential problems in genomic datasets” showcasing all the methodological development we are doing in this project such as the new nRCFV score. BOSTin itself is a tool to allow a user-friendly implementation of all these tools into a single one-stop solution. It is always fascinating to see how engaging he can present a complicated mathematical or bioinformatic subject for biologist.
Finally, our PhD student Alberto Valero Gracia presented “Homeotic genes and 3D genomics in Nemertea”. He gave an update on his work on the five Nemertean genomes generated with very high quality in his thesis. It also closed the circle to the first talk by Pia as it presented also the insights one can gain to look at the macrosynteny and microsynteny of these genomes. This time not from a phylogenetic perspective but from one on the evolution of these genomes.
All talks were very well received and resulted in lots of discussion afterwards. CEG could show that high-level genomic research is ongoing at the Natural History Museum and that we are involved in many genomic initiatives.
![]()

1 Comment on “CEG with a strong commitment to Norwegian Biodiversity Genomics”