As a proof of principle the European Reference Genome Atlas (ERGA) consortium initiated at the beginning a pilot study. Several sequences centers and different research projects contributed to this pilot study allowing the first sequencing of reference genomes across Europe and setting the stage for application of the Biodiversity Genomic Europe (BGE) project. With our InvertOmics project, we also contributed to this pilot study by sequencing the reference genome of the nemertean Emplectonema gracilis, a green slime worm. While we are still working on the release of this reference genome, some of us took a closer look at the selected species for the pilot study, how they were selected and especially the process from the sampling of species to the submission of the sample to sequencing center including different standards and their importance. We also provide an outlook to the future. The paper is now released as a preprint in bioRxiv and submitted for publication.
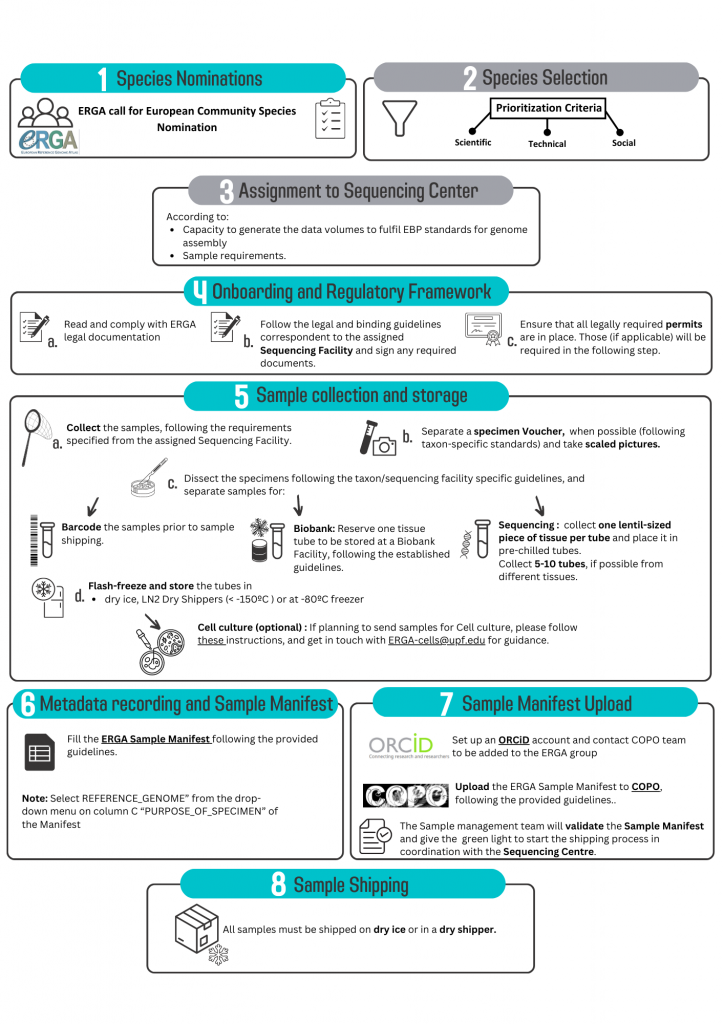
To support the generation of reference genomes for European biodiversity, the ERGA Sampling and Sample Processing committee (SSP) was formed by volunteer experts from ERGA’s member base. SSP aims to aid participating researchers through i) establishing standards for and collecting of sample/ specimen metadata; ii) prioritization of species for genome sequencing; and iii) development of taxon-specific collection guidelines including logistics support. SSP serves as the sample provider’s entry point to the ERGA genomic resource production infrastructure and guarantees that ERGA’s high-quality standards are upheld throughout sample collection and processing. With the volume of researchers, projects, consortia, and organizations with interests in genomics resources expanding, this manuscript shares important experiences and lessons learned during the development of standardized operational procedures and sample provider support.
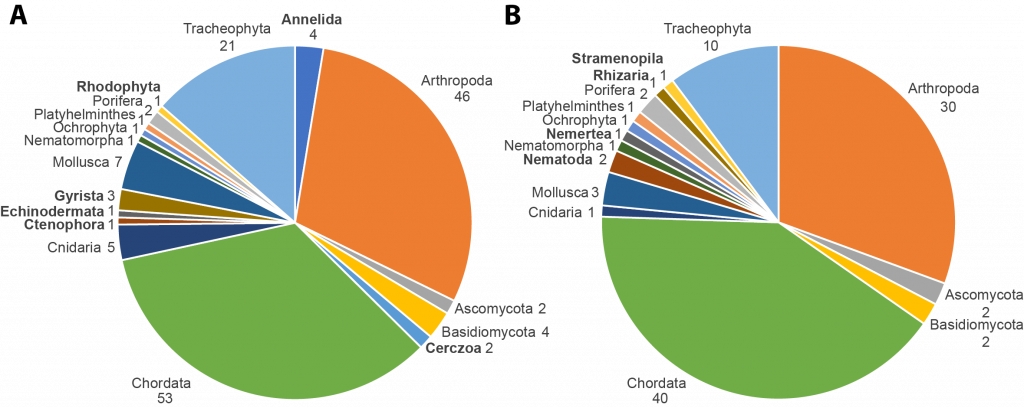
The manuscript details our experiences in incorporating the FAIR and CARE principles, species prioritization, and workflow development, which could be useful to individuals as well as other initiatives. For example, the pilot study comprised 98 species from 15 phyla and 34 countries or regions. Both the initial and the final list of selected species showed a predominance of chordates, arthropods, and plants. With six of the seven selection scores relating to feasibility, while the other criteria (i.e., conservation status, scientific relevance, socioeconomic relevance, taxonomic gaps, and community engagement) played only an indirect role. This as well as suggestions by the ERGA members may reflect the organism-bias of the biodiversity genomics community at large. Hence, this is still a major challenge towards providing a genome of each eukaryotic species on Earth even in a resourceful research community like the European one. A total of 37% of the species were considered for the category endangered/iconic, and 12% were pollinators (as one example of scientific relevance and a target group of the Biodiversity Strategy of the European Commission). The experiences of the pilot project are contributed to our development of the species prioritization processes in the community sampling process of BGE.
![]()

1 Comment on “ERGA pilot project – the selected species”