The answer to this question may lay in the paper I am presenting today and at some level genomes seem to be very stable. It was probably one of the most discussed papers at the museum this year. It was the topic of several journal clubs and shows how interesting and intriguing its findings. The paper by Simakov et al. (2022) “Deeply conserved synteny and the evolution of metazoan chromosomes” published in Science Advances had some very interesting results. The authors started from the finding that despite the known duplications of the whole genome and several rearrangements still relics of ancient bilaterian syntenies can be detected in vertebrate chromosomes. Synteny describes the presence of conserved blocks of genes of two species, which can be found on a chromosome together. Two different cases of synteny are considered: 1) microsynteny – containing a few genes, which have the same order in both species; 2) macrosynteny – the genes are on the same chromosome, but not in the same order. In this paper, the authors investigated the macrosynteny across the animal tree of life including a bivalve, a chordate, a sponge and two cnidarians.
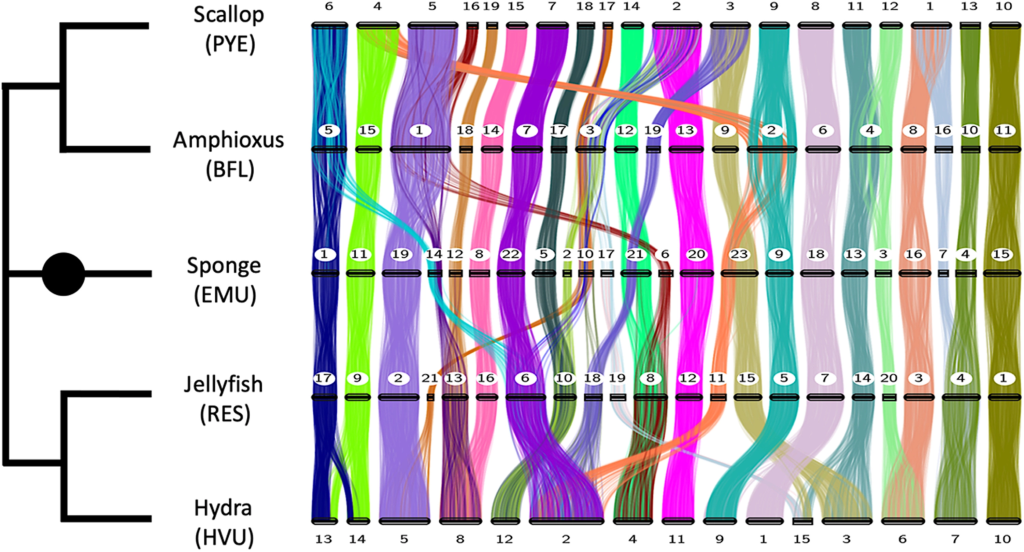
In their analyses, the authors can detect 29 ancestral linkage groups. The linkage groups probably reflect ancestral genomes, but this is not entirely certain and some might also represent the two arms of a chromosome. As the figure 1 shows, some of these linkage groups can be easily traced through time. For example, take a look at the chromosomes to the very right. Hence, some chromosomes can be very stable through time and conserved blocks of genes stayed together for more than half a billion of years. The authors also explored this stability further into the different animal groups such as lophotrochozoans, ecdysozoans, vertebrates and cnidarians. They found that the linkage groups as stay relatively stable even with increased numbers of species within the groups, but in some groups like the ecdysozoans is there also substantially more variability in the linkage groups.
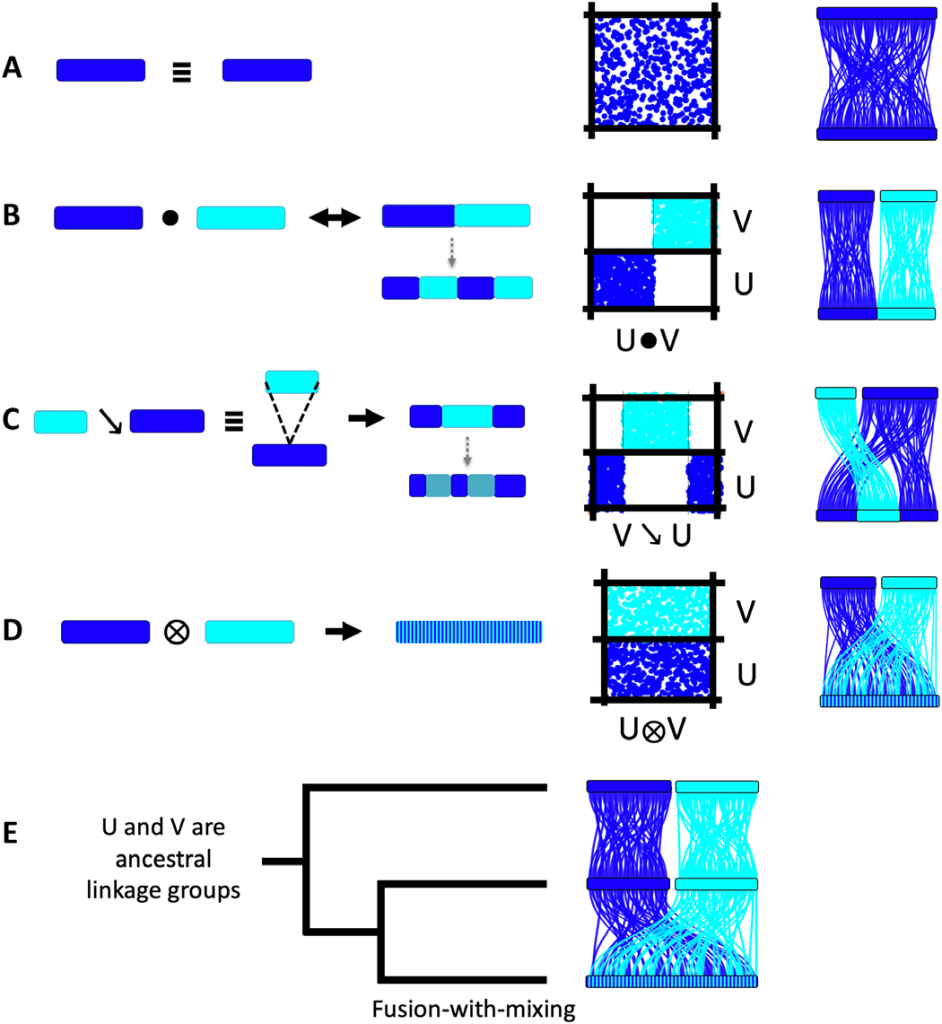
Furthermore, the authors investigated, which processes could lead to changes in the linkage groups. Besides the two already proposed processes, Robertsonian translocation and centric insertion, the author detected another process, which the authors called Fusion-with-mixing. This process starts with one of the two other processes and then they get mixed. This process is irreversible, might is also the most often found process in the paper, especially considering the processes happening at the basal branches of the trees.
The authors state in the first sentence of introduction: “The great variability of chromosome number across animals naively suggests that interchromosomal rearrangements have occurred frequently over the course of metazoan evolution”. This paper and others show that actually the chromosomes or linkage groups can be relatively stable over hundreds of millions of years. The same linkage groups can occur in sponge and chordate species and some smaller linkage groups are also found in the closely related unicellular eukaryotes. Why are these linkage groups are so stable?
The authors discuss two options here. “First, chromosome rearrangements may be constrained by gene regulation since structural variations that disrupt cis-regulatory interactions will generally be selected against.” However, the authors regard this as very likely as interchromosomal rearrangements occur at high frequency and would also disrupt cis-regulatory interactions. Second, “new chromosome-scale mutations must successfully navigate meiosis as heterozygotes on their way to fixation in a sexual population”. This scenario regard the authors as much more plausible. However, it is clear that much more work is needed to understand the degree of this kind of long chromosomal stability in the animal kingdom and which selective forces drive this stability. With this aspect of stability, it matches our research interests at the newly established STADIS (Stability & Discontinuity) hub perfectly.
Simakov, O., J. Bredeson, K. Berkoff, F. Marletaz, T. Mitros, D. T. Schultz, B. L. O’Connell, P. Dear, D. E. Martinez, R. E. Steele, R. E. Green, C. N. David and D. S. Rokhsar (2022). “Deeply conserved synteny and the evolution of metazoan chromosomes.” Science Advances 8(5): eabi5884.
![]()
