Our calendar is coming to its close and at the second-to-last day it features two Master projects, which started this year working with annelids, which are both completely computer-based making use of the SAGA supercomputer infrastructure through the command line, and all analytical programs used are monitored through coding.
The first project “How old are these worms? – Molecular dating of the annelid tree of life” is done by Stian Aleksander Helsem. He is a student in the Master program in Biodiversity and Systematics at UiO, after having completed the bachelor’s program in molecular biology (now Biovitenskap) at the University of Oslo.
The goal of his project is to date the annelid tree of life, initially using 1997 genes, presumed orthologs, downloaded from OrthoDB, comprising both nucleotide and amino acid datasets. The software OrthoGraph will be used as a first step eliminating paralogs and retaining orthologus groups, exclusively. After this, MAFFT will be used to generate alignments and IQTree for constructing single gene trees from these alignments. The amino acid datasets will be the only datasets examined further after this. A second round of orthology assessment will be conducted to screen for remaining paralogs and determining outlier sequences with extremely long branches due to contamination using TreSpEx. These steps will be performed in order to prune suspicious taxa from each gene. A second alignment and construction of single gene trees will then be performed on the pruned datasets.
Concatenation of the pruned genes will be done using FAS-con-CAT-G, and MARE will be used to remove uninformative genes from the supermatrix. Finally, different datasets will be generated containing for each family only the species with either the lowest degree of missing data, base composition heterogeneity or branch length. This will done both on a per-gene basis and also on the MARE reduced supermatrix as a whole. Each such compiled dataset will used to date the nodes in the annelid tree, using at least one Bayesian approach (mcmctree) and one Penalized Likelihood method (treePL). For this, he will use two different constraint trees based on recent literature and several fossil calibrations.
The second project “The meiofauna paradox and the degree of gene flow between supposedly sedentary animals” is conducted by Adel Abedpour Dehkordi. He is a Master student in the specialization Molecular biology and biochemistry of the Biosciences program at UiO. Before he obtained a Bachelor degree in Cellular and Molecular Biology from the Department of Biological Sciences of the Islamic Azad University (IAU) in Iran.
Meiofaunal or interstitial species are of small body size and/or inhabit the space between sand grains in marine sediments and soil. Many of these species are widely distributed, often over thousands of kilometers, contrasting with descriptions of a sedentary life style and the general absence of pelagic dispersal stages. These inconsistencies of a very wide distribution and the apparent lack of a dispersal stage became known as the “meiofauna paradox”. Hence, this raises questions how they can distribute this far. This question can be linked to the degree of gene flow between populations and species. Assessing the gene flow will help to understand their dispersal capacity.
Recently we identified a complex of several cryptic species of Stygocapitella in the World’s Ocean. To understand their evolution better we have obtained genomic level data to analyze these populations using double-digest restriction site-associated DNA-Seq (ddRAD). In this Master project, these already available data shall be used to investigate the gene flow within and between four Stygocapitella species occurring in the Northern Atlantic Ocean. The assessment of gene flow allows to assess the migration pattern and the degree of migration between the populations within a species and hence their dispersal potential. Moreover, as these species also occur in areas formerly entirely covered by glaciers during the last glaciations these studies will also shed light on how meiofaunal species reacted to the retreating ice after the last maximum and how fast they were able to colonize new areas.
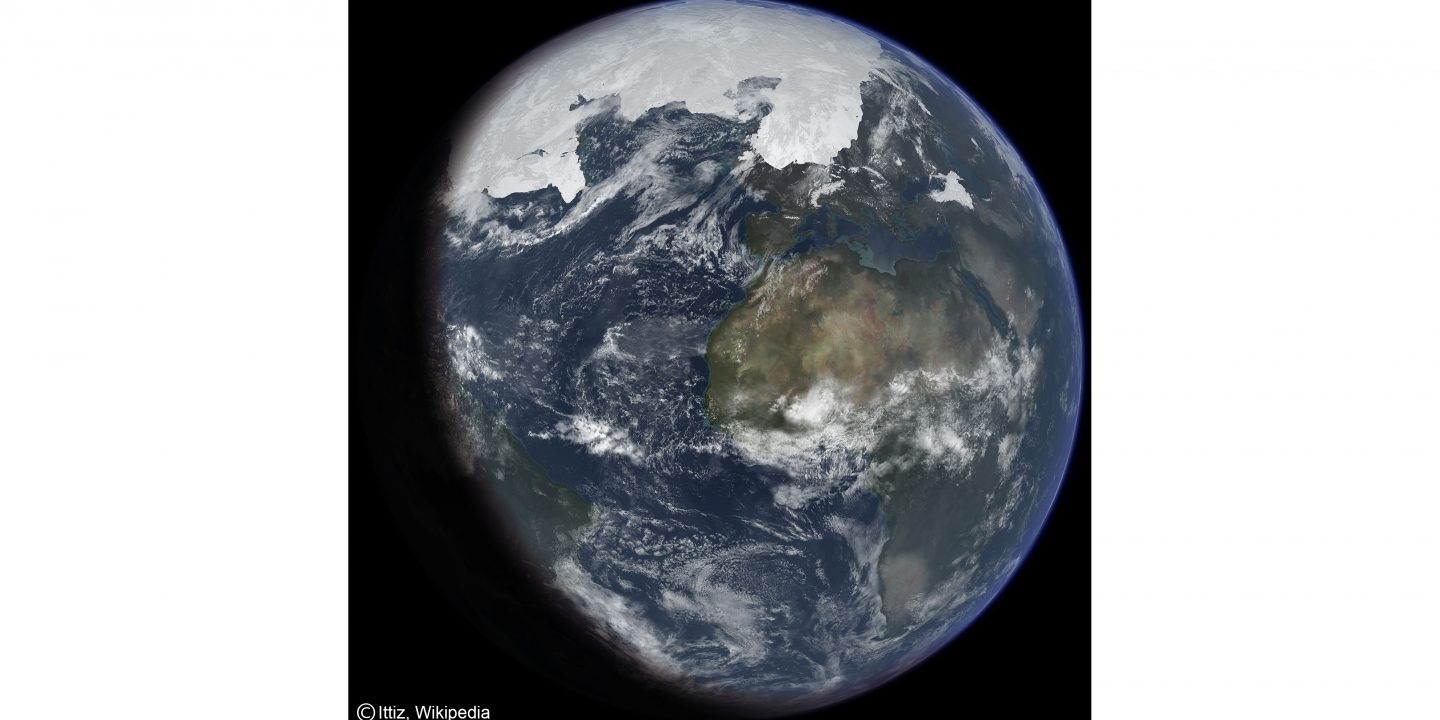
The featured picture above shows an artist’s impression of the last glacial period at glacial maximum.

Both pictures are changed to include the copyright, the used licenses can be found here (https://creativecommons.org/licenses/by-sa/3.0/deed.en)
![]()
1 Comment on “A tale of stone and ice”