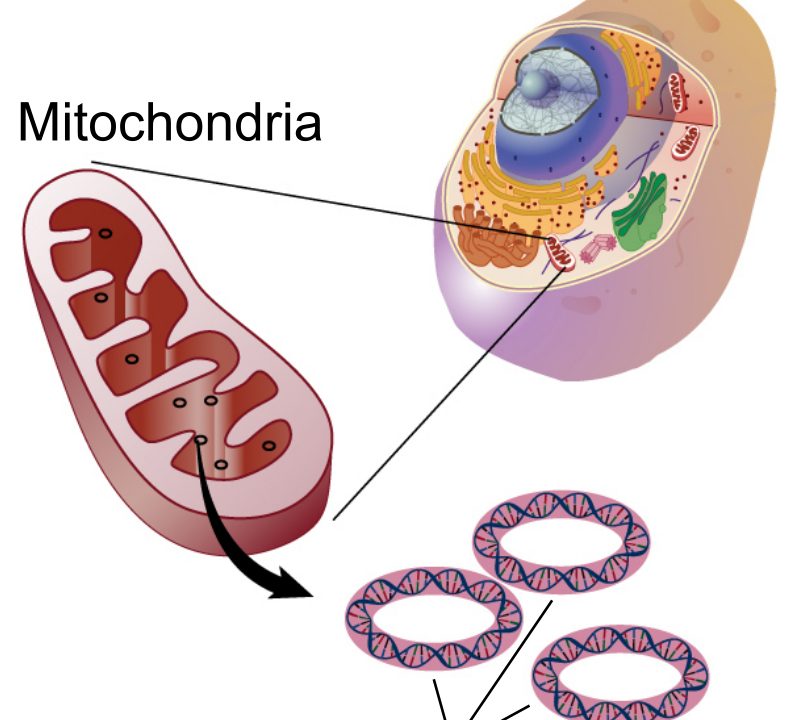
The mitochondria is the organelle within the cell which is regarded as the cells “power house”. This is because they play a crucial role in energy production through oxidative phosphorylation. Within the mitochondria, the mitochondrial genomes, also referred to as mitogenomes, are found. The mitogenomes are small circular DNA molecules, often in the size range from 14kb to 20 kb in the majority of animals. The mitogenomes are referred to in plural, because each cell can contain multiple copies of the mitochondrial genome.
The mitochondrial genome has multiple interesting and favorable characteristics making them very interesting for us working in different fields of biology including evolutionary studies, population genetics and medical research to mention some.
Characteristics of the mitochondrial genome
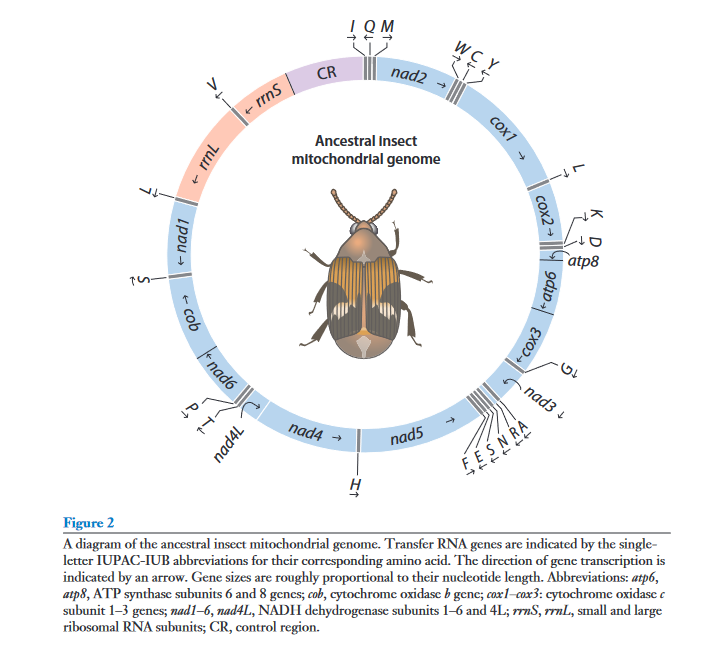
- Circular DNA: Mitochondrial genomes typically consist of a closed, circular DNA molecule. This circular structure is a common feature across a wide range of organisms, including animals, plants, fungi, and protists.
- Maternal Inheritance: One distinctive feature of mitochondrial genomes is their mode of inheritance. In most cases, mitochondria, and consequently their genomes, are passed from the mother to the offspring. This maternal inheritance pattern is a result of the fact that the mitochondria in the sperm are often excluded during fertilization, and thus, the mitochondria in the offspring are derived exclusively from the maternal side.
- Gene Content: Despite their relatively small size, mitochondrial genomes typically encode a set of essential genes involved in oxidative phosphorylation and energy production. These genes include those that encode components of the electron transport chain and ATP synthesis machinery. The specific genes present can vary between different organisms, but common elements include genes for cytochrome c oxidase, NADH dehydrogenase, and ATP synthase.
- Coding Regions and Non-Coding Regions: The mitochondrial genome consists of both coding and non-coding regions. Coding regions are responsible for producing functional proteins, whereas non-coding regions include intergenic spacers and regions involved in the regulation of mitochondrial gene expression.
- High Copy Number: Each cell can contain multiple copies of the mitochondrial genome, and the number can vary depending on the cell type and energy requirements of the tissue. This high copy number helps ensure an adequate supply of mitochondrial DNA for cellular functions.
- High mutation rate: useful for examining recent evolutionary events and fine-scale population dynamics. In species with short generation times, such as many invertebrates, plants, and certain vertebrates, mitochondrial genomes can be employed to investigate population structure, migration patterns, and demographic history. The rapid evolution of mitochondrial DNA is particularly advantageous for resolving relationships among closely related taxa.
Understanding the structure and organization of mitochondrial genomes is as mentioned important for various fields of biology. Mitochondrial genomes play a crucial role in the field of systematic biology, providing valuable insights into the evolutionary history, phylogenetics, and population genetics of diverse organisms. Especially using the mitochondrial genome for phylogenetic research, is something our colleagues and we at the museum are commonly doing a lot of.
Mitochondrial DNA in phylogenetic work
Mitochondrial DNA has been considered to be a fairly reliable marker in phylogeny of both closely and distantly related insect taxa as well as animals in general (Stephen L. Cameron, 2014; Stephen L Cameron, Lambkin, Barker, & Whiting, 2007) (and references therein). Mostly because of its technical ease-of-use, maternal inheritance, near-neutrality, lack of recombination, and rapid evolutionary rate compared with nuclear DNA (Dong, Wang, Li, Li, & Men, 2021). A very common mitochondrial gene, or marker, to use, is Cytochrome c oxidase I, which is widely used and is found to be the most conserved gene in terms of amino acid evolution among the protein-coding genes (Simon et al., 1994). Cytochrome c oxidase I (COX1, COI or CO1) also known as mitochondrially encoded cytochrome c oxidase I (MT-CO1) is a protein that is encoded by the MT-CO1 gene in eukaryotes, is also the preferred barcode marker for most animal species.
Conserved structure and high mutation rate
One of the key features that makes mitochondrial genomes significant in systematic biology is their relatively conserved structure across many eukaryotic species. This allows for the identification of homologous regions and the comparison of sequences between different taxa. Mitochondrial DNA sequences, particularly those encoding essential functional genes, can serve as molecular markers for studying evolutionary relationships and constructing phylogenetic trees. Further, the high mutation rates observed in mitochondrial DNA make it especially useful for examining recent evolutionary events and fine-scale population dynamics. In species with short generation times, such as many invertebrates, plants, and certain vertebrates, mitochondrial genomes can be employed to investigate population structure, migration patterns, and demographic history. The rapid evolution of mitochondrial DNA is particularly advantageous for resolving relationships among closely related taxa.
However, it’s crucial to note that the exclusive reliance on mitochondrial genomes for evolutionary studies has limitations, and researchers often integrate data from nuclear genomes and other molecular markers to obtain a more comprehensive understanding of evolutionary processes.
Recent use of mitochondrial data by the FEZ group
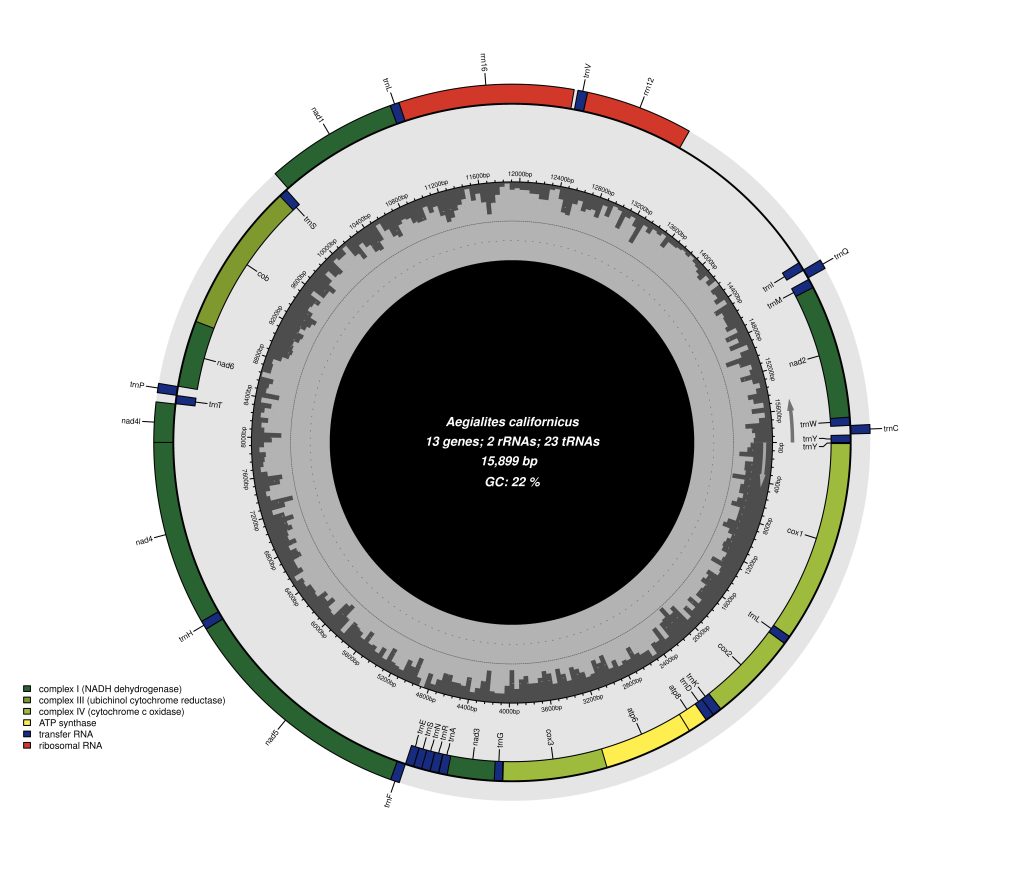
Since mitochondrial is very commonly used both as a barcode marker and for phylogenetic studies, the majority of phylogenetic studies include at least one mitochondrial gene (and preferably one nuclear gene). As an example, the first project of my PhD is reliant on a phylogenetic tree of the genus Aegialites using the mitochondrial marker COI and the nuclear ITS2 marker. Having a phylogenetic backbone of your desired study group, allows you to get an overview of e.g. number of species in a genus and how they are related to each other. With such a phylogenetic tree one can make a stronger inference of the direction of your next research questions and you would know which species to include in your next study in other areas like population genetics, hybridization, dispersal, trait evolution and so on. Further, our group has also produced some interesting papers using mitochondrial DNA the last couple of years you can have a look at:
Mitochondrial Genome Evolution in Annelida – A Systematic Study on Conservative and Variable Gene Orders and the Factors Influencing its Evolution. Struck, T. H., Golombek, A., Hoesel, C., Dimitrov, D. & Elgetany, A. H. (2023) Systematic Biology 72: 925–945. https://doi.org/10.1093/sysbio/syad023
Mitogenomics and the genetic differentiation of contemporary Balaena mysticetus (Cetacea) from Svalbard. Bachmann, L., Cabrera, A. A., Heide-Jørgensen, M. P., Shpak, O. V., Lydersen, C., Wiig, Ø., & Kovacs, K. M. (2021). Zoological Journal of the Linnean Society, 191(4), 1192-1203.
Complete mitochondrial genome of the Galápagos sea lion, Zalophus wollebaeki (Carnivora, Otariidae): paratype specimen confirms separate species status Austin R.M., Eriksen, P.M. & Bachmann, L. ZooKeys 1166: 307-313. https://doi.org/10.3897/zookeys.1166.103247
References
Cameron, S. L. (2014). Insect mitochondrial genomics: implications for evolution and phylogeny. Annual Review of Entomology, 59(1), 95-117. doi:10.1146/annurev-ento-011613-162007
Cameron, S. L., Lambkin, C. L., Barker, S. C., & Whiting, M. F. (2007). A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Systematic Entomology, 32(1), 40-59.
Dong, Z., Wang, Y., Li, C., Li, L., & Men, X. (2021). Mitochondrial DNA as a Molecular Marker in Insect Ecology: Current Status and Future Prospects. Annals of the Entomological Society of America, 114(4), 470-476. doi:10.1093/aesa/saab020
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H., & Flook, P. (1994). Evolution, Weighting, and Phylogenetic Utility of Mitochondrial Gene Sequences and a Compilation of Conserved Polymerase Chain Reaction Primers. Annals of the Entomological Society of America, 87(6), 651-701. doi:10.1093/aesa/87.6.651
![]()