The bowhead whales (Balaena mysticetus) in the East Greenland Sea, the Svalbard region and the Barents Sea (referred to as the East Greenland-Svalbard-Barents Sea (EGSB) stock) has been intensively studied for many years by members of the FEZ group. The stock is classified as Endangered in the International Union of Conservation of Nature (IUCN) Red List after it has been hunted to near extinction. The size of the stock has been discussed for years, and also the impact of the extensive hunting on the genetic diversity has been of concern.
An international research team led by FEZ scientists has now published a study in the prestigious scientific journal Scientific Reports addressing the genetic diversity of EGSB bowhead whales. The genomes of twelve bowhead whale individuals sampled in 2017 and 2018 were sequenced by next-generation sequencing. The paper provides important baseline data to further evaluate the impacts of various potential stressors such as environmental change on this stock.
The authors report a relatively high level of genetic/genomic diversity; there was no indication of significant inbreeding and a mean heterozygosity three times higher than a conspecific individual sampled in West Greenland. Mean heterozygosity is a measure indicating how many positions in a genome show different nucleotides. High heterozygosity means lots of genetic variability, and low heterozygosity means little genetic variability. That the EGSB bowhead whales retained relatively high genetic diversity despite the intense hunting may nevertheless not be too surprising. Bowhead whales are long-lived animals that can become older than 200 years, and, accordingly, only few generations have passed. Even if hunting was extreme, the population size would have needed to be very small over a long period to note substantial reductions in genetic diversity in the present. However, if the small population size continues, a rapid accumulation of inbreeding is to be expected.
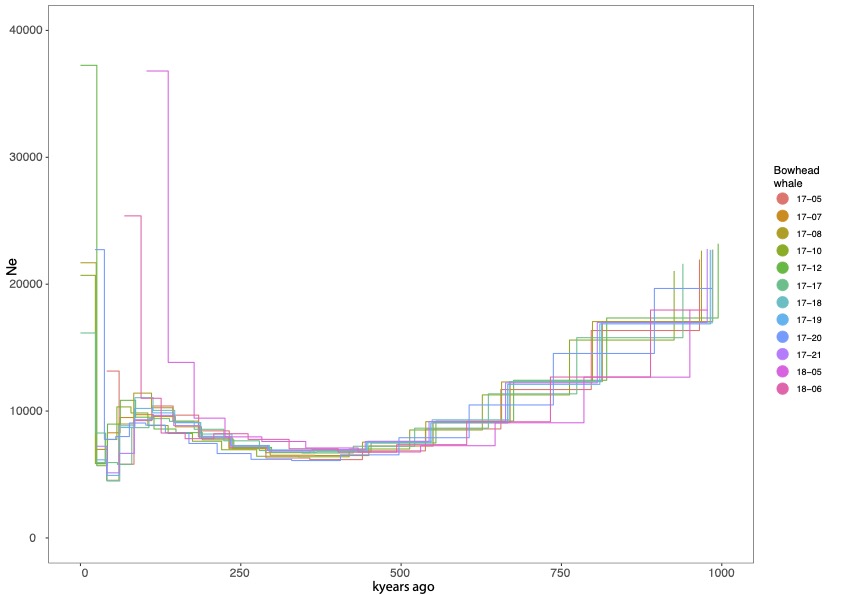
The information of genetic variability of the individual genomes also allowed for modelling the demographic history of the EGSB stock. There was a rather constant decrease in the effective population size (Ne) from roughly 30 000 individuals 1.5 million years ago to ca. 7000 individuals about 250 000 years ago, which was followed by a slight increase up to ca. 9000 at 75 000 to 100 000 years ago. Roughly speaking,, the effective population size (Ne) is the number of breeding individuals in the population, and the the census population size is usually larger. The population size estimates were lower than those reported earlier by Bachmann et al. (2021) based on the analysis of mitochondrial genomes of the same bowhead whale individuals. However, mitochondrial diversity is not always nicely correlated with population size, social structure and matrilineal social systems may be important drivers of range-wide and regional mitochondrial genetic diversity.
The authors of the new study emphasise that they provide first nuclear genomic baseline data for the EGSB bowhead whale stock. They call for more comprehensive analyses of genetic differentiation and for the reconstruction of the demographic history of the other bowhead whale stocks in the future.
References:
Cerca, J., Westbury, M.V., Heide-Jørgensen, M.P. et al. High genomic diversity in the endangered East Greenland Svalbard Barents Sea stock of bowhead whales (Balaena mysticetus). Scientific Reports 12, 6118 (2022). https://doi.org/10.1038/s41598-022-09868-5
Bachmann, L., Cabrera, A.A., Heide-Jørgensen, M.P.et al. Mitogenomics and the genetic differentiation of contemporary Balaena mysticetus (Cetacea) from Svalbard. Zoological Journal of the Linnean Society 191, 1192–1203 (2021). https://doi.org/10.1093/zoolinnean/zlaa082
![]()

1 Comment on “Genomic Diversity of endangered bowhead whales”