For the 6th day of our Journal Review Advent Calendar, I wanted to present Jesus Lozano-Fernandez’s brilliant “A Practical Guide to Design and Assess a Phylogenetic Study”. I think reviews like this are really important – whether you’re trying to grasp a new kind of analysis for research, studying for an exam, or really need to understand the methods of another paper, comprehensive, introductory reviews are the perfect place to start.
https://academic.oup.com/gbe/article/14/9/evac129/6659225
Phylogenetics is a broad field, but more and more studies in biological journals want to see phylogenies inside as many studies as possible. I think there are a lot of good arguments both for and against this practice, but with the trend being what it is, it’s important that when it is done, it’s done well. Lozano-Fernandez begins with an extensive, but comprehensive introductory glossary: phylogenetics has a broad library of technical jargon, and I think the explanations here really help demystify the whole exercise and make it easy to sift out the Incomplete Lineage Sortings from the Hidden Paralogies.
Following a brief introduction of the history of gene sequencing and the genomic revolution, Lozano-Fernandez then dives into the meat of the review, immediately setting out the reasons why errors occur in phylogenetic studies, and trying to expound on the reasoning behind these. Clearly splitting up error caused by biology, and errors caused by model violation helps the reader to centre their own data and their own understanding of biology inside the world of phylogenetics.
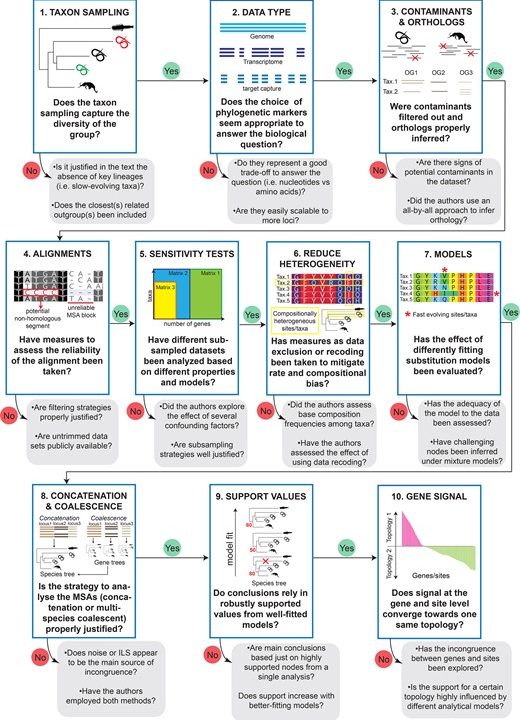
Although the review is not partitioned into separate bookmarked sections (Just an Introduction and Conclusion), it then transitions into a step by step guide of a phylogenetic study, leading the reader from dataset selection through alignment and sensitivity testing, until the phylogeny construction and final result. This is all very nicely and clearly shown by the main figure of the publication, that is also the featured image! Explaining different issues that might arise with DNA and RNA, nucleotide and amino acid data, it is a brief but well-structured introduction to the practice of phylogenetic reconstruction.
Covering such a broad topic in such a brief space – the review comes in at only 10,000 or so words – is a difficult ask, and there are definitely areas that, if you’re having issues with a particular dataset, you might want to look at more closely. Dataset Selection and Artifact Identification, mentioned in the introduction, only really have space for a few thousand words that fairly distills topics with over 50 years of data. That said, this review is clearly intended as a first step for new practitioners, and I think for its goal, it really hits the back of the net. I’ve been sending it on to colleagues and new students as they start to familiarise themselves with these kind of analyses, and now I’m shamelessly promoting it on the blog here too. If you’re in need of an accessible intro to phylogenetics, look no further!
![]()