This year the FEZ group got a new FRIPRO project funded by the NFR called “InvertOmics – Phylogeny and evolution of lophotrochozoan invertebrates based on genomic data”. The project includes many members from FEZ (Alberto, James, Lutz and Torsten), other researchers at the NHM and international collaborators. So what is the project about and what will we do the next years?
How the last common ancestor of animals like humans, fishes, insects, roundworms, snails and earthworms looked and how evolution proceeded within this large group of animals is discussed in biology. One hypothesis suggests that evolution advanced from a simple organization similar to flatworms towards more complex forms several times independently, while another one favors the opposite direction from a complex, bristle worm-like ancestor to simple organizations by several separate reductions. Support for one or the other hypothesis depends on how the phyla in the group Lophotrochozoa, comprises 16 of the 32 animal phyla, are related to each other. This is presently unknown, and one of the major reasons for this are that only very few genomes have been sequenced for this group, and that the genomic data are heavily affected biases, which can mislead analyses trying to resolve their relationships.
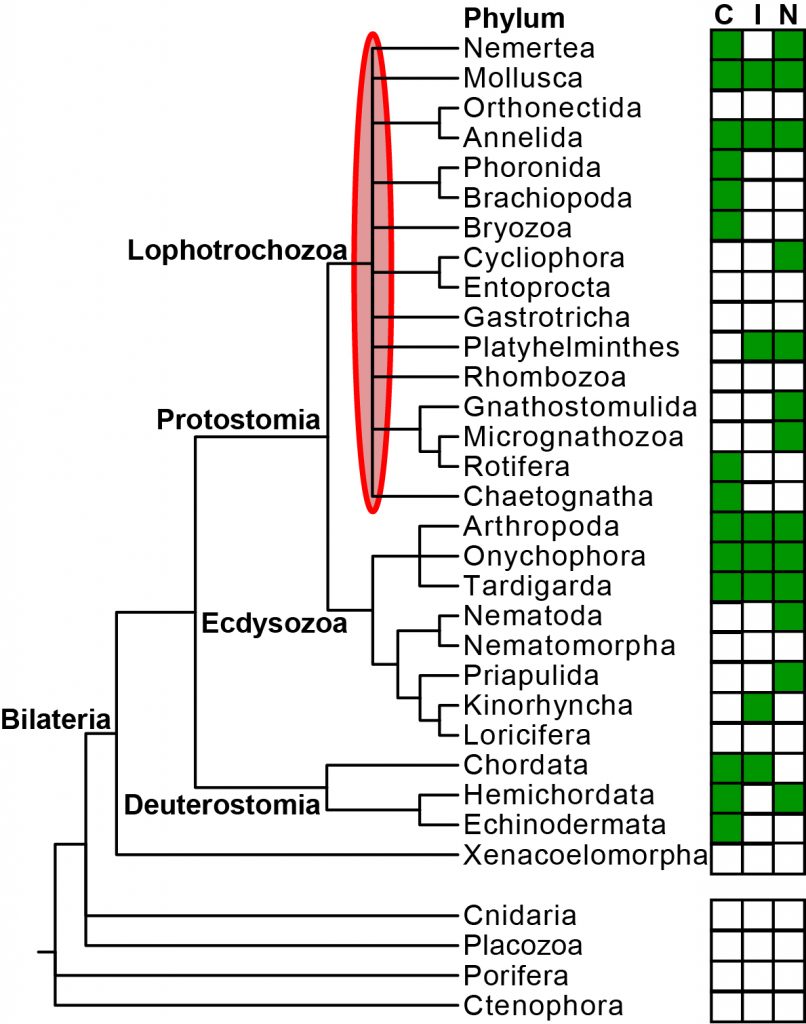
In the InvertOmics project, we aim to generate high-quality genomes for 50 species covering all lophotrochozoan phyla using modern sequencing technology. Several lophotrochozoan species have very small body sizes (smaller than 0.5 mm), but we will address this challenge by adapting existing single-cell genomic protocols from cancer research. High-quality genomes will be determined for the first time for such species. We will also develop new bioinformatic tools. They will allow us to ameliorate the effects of the misleading biases. Moreover, this will also include a new support measurement, which is entirely different from all recent measurements. Due to both these new genomes and tools, we will be able find out how the different lophotrochozoan phyla are related to each other and how the last common ancestor of animals like humans, insects, and earthworms looked and how evolution proceeded within this large group. An additional of all these genomes is that they provide new opportunities to study genome evolution, developmental pathways, ecology or to discover new medicinal drugs.
Text in the teaser: Want to learn a bit more about InvertOmics and the figure means, look at the first door of the advent calendar (December 1st).
Picture in the teaser: Same as above.
![]()
2 Comments on “The first door of the calendar”