Pia Merete Eriksen, Rita M. Austin
In June 2021, I, Pia M Eriksen, conducted a research project under the guidance of Rita M Austin to sequence herptile type specimen DNA using in-house sequencing techniques, supported by Oslo Natural History Museum and UiO: Life Sciences. Now, 5 months later, we have concluded the wet-lab and sequencing portion, and I have returned with our findings.
Briefly, we aimed to test the impact of age and formalin on DNA yield and quality from herptile, tapeworm, and hooded seal Oslo NHM museum specimens when using an in-house, cutting-edge sequencing technology (Oxford Nanopore MinION Mk1C). Collected between 1855 and 2008, and with various degrees of formalin fixation, we assessed tissues from six species and individuals: the >100 year old and low-formalin treated herptile type specimens Ranoidea dahlii, Lipopholis kintorei, Ctenotus pantherinus, and Ctenotus colletti; Diphyllobothrium latum, the >50 year old, high-formalin treated tapeworm; and the no-formalin, modern hooded seal (Cystophora cristata) (Figure 1).
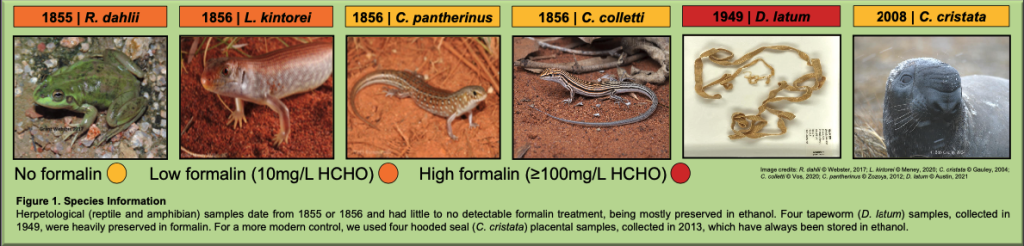
Herpetological (reptile and amphibian) samples date from 1855 or 1856 and had little to no detectable formalin treatment, being mostly preserved in ethanol. Four tapeworm (D. latum) samples, collected in 1949, were heavily preserved in formalin. For a more modern control, we used four hooded seal (C. cristata) placental samples, collected in 2008, which have always been stored in ethanol.
We extracted DNA following a multi-day incubation protocol optimised for formalin-fixed tissues (Gould, Fritts-Penniman and Gaisiner, 2021) and using the QIAmp DNA Micro Kit. After estimating DNA fragment length and measuring DNA concentration, we attached sequencing barcodes and adapters; think of the barcodes as a library’s Dewey Decimal code and the adapters as the hardcover book binding. Following sequencing, data was quality filtered, and barcodes and adapters were removed (to only assessed the species DNA). Analyses compared yields of DNA pre and post sequencing, and the impact of quality filtration.
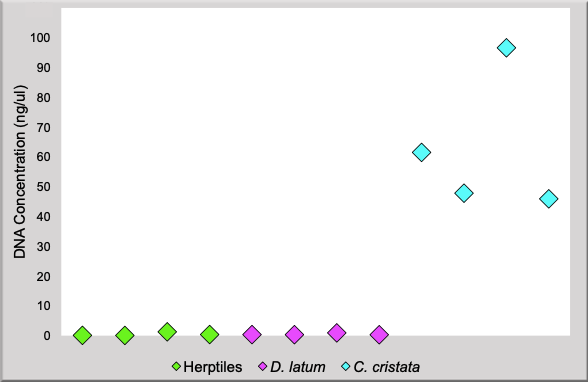
The concentrations of DNA for the aged herptiles and formalin fixed D. latum samples do not exceed 0.1ng/uL, whilst for C. cristata, concentrations are well above 40ng/ul. This indicates that significantly more DNA was accessible from the modern, ethanol treated C. cristata samples. This illustrates the impact of age and preservation method on the accessibility of DNA.
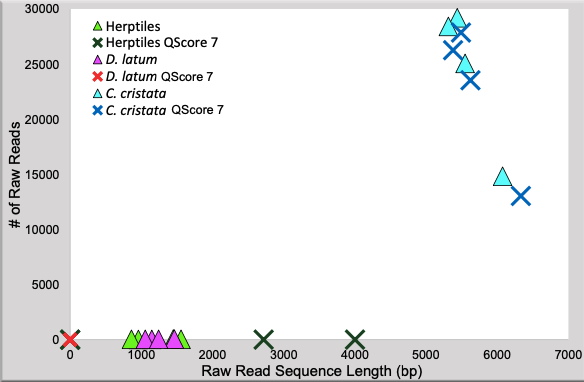
A similar amount of DNA was initially sequenced from the D. latum and herptile specimens. Post base quality filtration, no sequences remained for D. latum samples and two herptile individuals (C. colletti and R. dahlii). Only five and six passing sequences remained for L. kintorei and C. pantherinus, respectively. C. cristata samples yielded a higher number of sequences (~13000 – 27000 per sample) and longer reads, both before and after quality filtration. This implies that the presence of formalin is more restrictive than age. Results indicate that the sequencing platform is only reliable for specimens with long and abundant DNA fragments
Our study contributes to understanding the accessibility of museum collection DNA using the Nanopore MinION Mk1C platform. We found that formalin was used in varying intensities across museum collections and taxon, pointing to the arbitrary decision makings of previous collectors and curators. DNA concentration is heavily impacted by formalin concentration and age. The MinION Mk1C platform functions optimally with modern samples that are neither fragmented by age nor cross-linked by the application of formalin treatment. Post quality filtration, only longer sequences are kept, which greatly reduced the number of analysis ready sequences. Only C. cristata sequences yielded species informative data.
This study sparks questions on a broader scale, such as “how should museums proceed with destructive sampling decisions, given the likely application of formalin in wet-collections?”. This is an on-going concern due to the finite number of historic specimens and tissues available in museum collections. Limitations for this study include human error (e.g. dilution inaccuracies during final sample pooling), limited sample size, and the the MinION Mk1C platform’s preference for long sequences.
Future studies should consider the impact of age and chemical treatment when sequencing DNA from museum specimens. Beneficial follow up experiments could look at other chemical treatments to correct and/or mitigate the effects of crosslinking, as well as investigating other sequencing platforms that may be more suited to shorter fragments.
Reference: Gould, A.L., Fritts-Penniman, A. and Gaisiner, A. (2021) ‘Museum Genomics Illuminate the High Specificity of a Bioluminescent Symbiosis for a Genus of Reef Fish’, Frontiers in Ecology and Evolution, 9. doi:10.3389/fevo.2021.630207.
![]()
