Author: Rita Austin (former group member)
New research highlights and tests the limitations of dental calculus (i.e., calcified dental plaque), a microbiome substrate regularly used to reconstruct ancient foodways. Under the direction of Allison E. Mann from Clemson University, a collaborative team analyzed both synthetic and ancient dental calculus datasets to demonstrate the intrinsic challenges related to identifying, authenticating, and interpreting ancient microbiome dietary information.
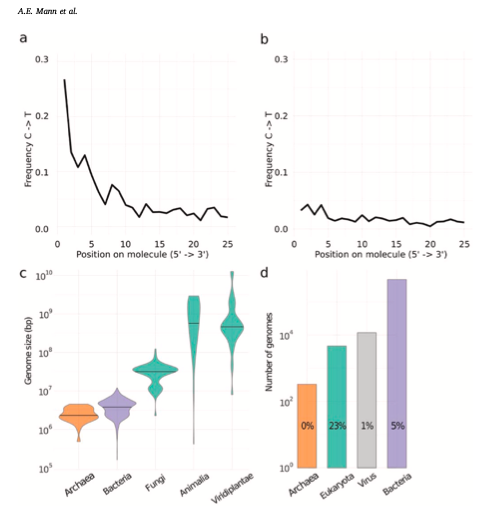
Essential to human culture, biology, and evolution, ancient diet reconstructions help us to understand a fundamental component of the human experience. Dental calculus contains host, microbial, and to a lesser extent, dietary biomolecules, making it an attractive and commonly used substrate for diet reconstruction using metagenomic techniques. However, several factors hinder the accurate detection of dietary biomolecules, including: (1) the fragmented and damaged nature of the DNA molecules, making sequence identification and authentication particularly challenging, (2) the relatively low number of dietary biomolecules as compared to those derived from the endogenous and environmental microbial community, (3) differences in genome size and similarity across different species, and (4) reference database errors or lack of representation.
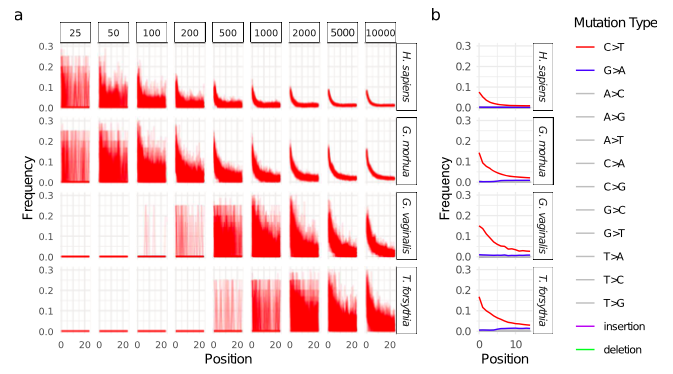
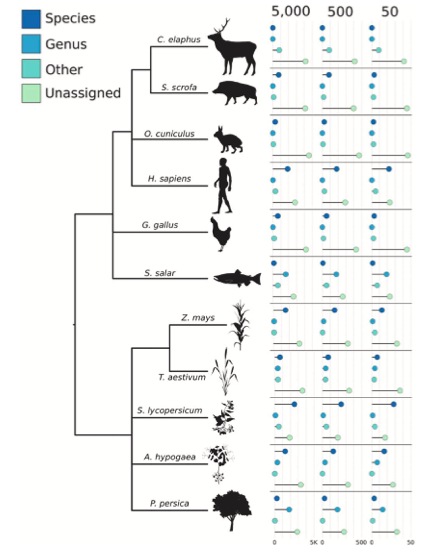
To assess the challenges intrinsic to genetic analysis of diet, various metagenomic communities were generated. Results emphasized the inherent challenges to current ancient diet reconstructions, calling for more stringent and consistently applied guidelines. Authentic damage patterns require at least ~500 reads for eukaryotes, with microbes needing thousands of reads (Figure 1 and 2). All of the dietary and host organismal reads incorporated into the synthetic datasets were not able to be assigned to any taxonomic level, with many regions across eukaryotic genomes mapping equally well to unrelated sources (Figure 3). Mapping mismatches direct dietary interpretations to organisms that ‘make sense’, relative to archaeological and temporal contexts. However, relying upon reads identified to organisms expected/suspected to be contextually relevant obstructs the greater complexity of eukaryotic results from metagenomic data and directs interpretations post hoc.
Importantly, expansion of eukaryotic genomic reference databases can help alleviate the challenges identified in the paper. With more, well-covered eukaryotic genomes, uninformative reads mapping to conserved genome regions can be pushed up to broader taxonomic levels through least common ancestor identification methods, as well as providing diverse comparative references. The authors list key evaluation criteria for assessing studies conducting dietary analyses from ancient dental calculus. In following the evaluation points, more study transparency can be gained, and the validity of study claims can be assessed.
Read the full results in: Mann, Allison E., James A. Fellows Yates, Zandra Fagernäs, Rita M. Austin, Elizabeth A. Nelson, and Courtney A. Hofman. “Do I have something in my teeth? The trouble with genetic analyses of diet from archaeological dental calculus.” Quaternary International (2020).
![]()