Last week, as part of a special issue of Zoologica Scripta, I started the year with my first paper – and my first solo effort! “The Wealth of Shared Resources: Improving molecular taxonomy using eDNA and public databases” is a rather portentous title for a pretty simple experiment. I looked at all of the entries for a certain type of gene sequence in a public gene database, and reidentified some of the older sequences with current knowledge. But all studies have a story, and this is that story.
The year was 2020, the month was December and, thanks to some negative Covid test results via a clinic in Tokyo, and a rushed change at Charles de Gaulle, I had just arrived in Oslo. In the middle of a lockdown. Thanks to the wonders of the internet, I’d managed to secure a flat before arriving, and after Torsten kindly picked me up at the airport, I found that my new landlady had bought me an oven pizza and a bottle of beer.
In this way, I had a chance to get my bearings before working out the simple things: like how to get food in a new country under lockdown, where you don’t know anybody, and are quarantined (the answer, by the way, is ordering groceries online, then loitering outside for the delivery because your flat doesn’t have a number and your doorbell doesn’t have your name on it yet. Not particularly efficient, but it works!).
Rather than confront those issues immediately, I instead flopped down onto the settee with a slice and a beer and started thinking about tardigrades. As I imagine everyone must do when faced with change.
More specifically, I started thinking about how, when I was working on another paper (that would get blogged about here), I was encountering a lot of tardigrade 18S sequences when looking through GenBank that weren’t assigned to any specific group of tardigrade. They were just listed as “tardigrade”. Were they the bulky, tank-like Milnesium? The iconic Ramazzottius, focus of so much tardigrade research? Tanarctus, with its eye-catching balloons? 18S sequences are commonly used to work out how different species are related to one another, and especially in a group with so few available genomic sequences, being able to update these definitions could be useful. And if we could prove that it worked – that method could have applications outside of tardigrades, to other undersampled or poorly understood groups, like the ones we work with in Invertomics!
And so, in the day I worked on my Lophotrochozoa with Torsten. And on cold winter nights, with nobody around to chat, I worked on this.

Genbank is a public resource of protein and DNA sequences that is freely available to access, and has been since the 90s. This means, in turn, that there is a heck of a lot of stuff on Genbank. And whether you’re sitting in a university, or just have spare time and a question, you can access it all. This idea of public, open access permeates this study too – every bit of software I used was freely available, and all of the bits that were computationally difficult to do were done on freely accessible web servers hosted by universities and research institutes across the globe. That means that I could have done it on a state of the art supercomputer, or a 20 year old laptop.
Working with eDNA though, had its own challenges. eDNA stands for “Environmental DNA”, and means that, rather than picking up a specimen, identifying it under a microscope (morphological identification), and then sequencing DNA from it, you get an environmental sample of earth, water, or air and sequence it all at once from micro-organisms to loose hair. The findings are then compared to databases we have of known DNA samples to see what might be living in the area. Since sequences might be made up of fragments of DNA from lots of different individuals, they aren’t very good for the taxonomic studies I normally do (working out which species are what, whether morphological identifications are correct or more complicated, and how different species are related to one another). But used alongside some of the sequences with good identifications, we might be able to work out, at least generally, what these unknown tardigrades are. And that might help us know where to look for certain kinds of tardigrades in the future, or reveal exciting and unknown things about tardigrades. And we can do it all for free!
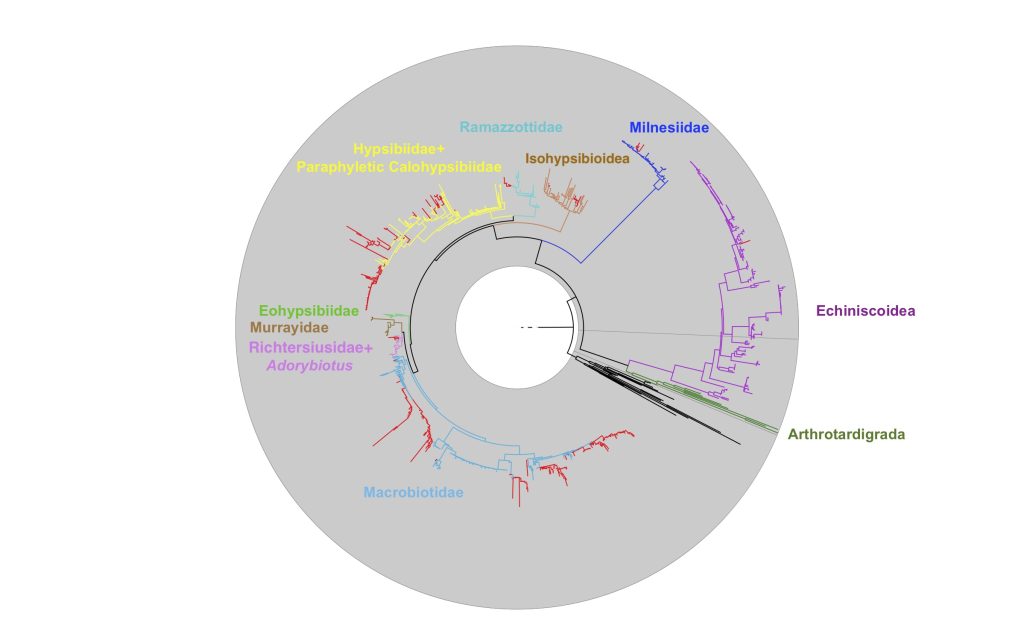
What I found is that, for a lot of these sequences, I was able to identify them down to family level. Tardigrades are a phylum – that means they are a classification at the same level as Chordates – animals with a backbone and the closely related friends of animals with a backbone (like Lancelets and Tunicates).
If I said “oh, I’ve got… a vertebrate sequence (Chordate)”, I think a fair reply would be “cool, which one?”. The family level, however, is the same as being able to say “I’ve got a cat (Felidae)”, but for tardigrades! It’s a pretty thrilling improvement. We weren’t able to make distinctions between tardigrade families back when these eDNA studies were first published in the 2000s, because we just didn’t have enough sequence data about them. But now we do – which I think also says a lot about the exciting progress we’ve made in the last few decades.
But that also implies that there are thousands more of these sequences in GenBank that might benefit from a similar second glance. With all of the advancements we have made, and all the extra samples we have now, we might look back and find that we can use the data we collected in the past in entirely new ways!
And from my comfy settee in Oslo, with pizza in hand (though a different settee since I moved and, thankfully, a different pizza from two years ago too), I think that’s pretty neat.
![]()