Usually I work with marine invertebrates from a group called Lophotrochozoa, which comprises among others mollusks, segmented worms and flatworms. However, the paper featured in this blog is the first of several papers to come on insects. While the others will come from Marianne’s PhD project on Aegialites beetles in our group (see her blog entries on her project), this one on the fungus gnats of the genus Allodia is a collaboration of three research groups at the NHM (Ento, FEZ and SERG) and lead by Trude Magnussen as part of her PhD project.
Fungus gnats are known from all continents except Antarctica and they have an overall uniform morphology allowing for that several genera can only be diagnosed by subtle differences in characters, especially in male genitalia. Allodia is one of the genera in this group and contains 96 described species in two subgenera. The genus is widely distributed occurring among others in Holarctic, Afrotropical, Oriental and Oceanian regions with the majority from the Holarctic. Species of Allodia are small to medium-sized with slender body and long legs and can be easily separated from other genera by several characters such as branches of both forks without setulae, wings with subcostal vein ending in vein radial vein and a thorax with a bare anepisternum and scutum with bristles, but not evenly clothed by setae (Fig. 1). The two subgenera of Allodia, i.e., Allodia and Brachycampta, were originally described as separate genera, but later they were merged to one genus Allodia. Characters to distinguish between the two subgenera exist, but are usually subtle.

Photo: Karsten Sund (NHM, Oslo).
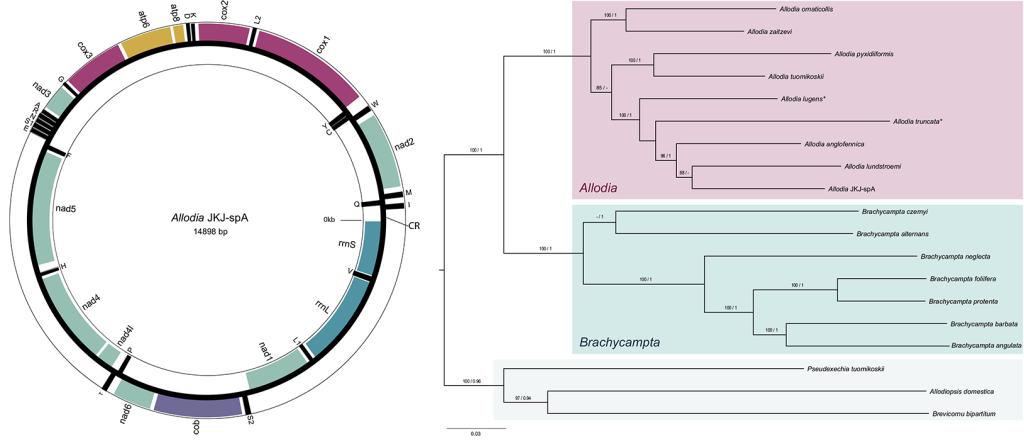
To test if it were two genera or one as presumed at present, we used a genome skimming approach in combination with further analysis of morphological characters. Genome skimming means the genome of an individual is sequenced only at a low coverage of less than 10x. This means on average each position of the genome is sequenced less than 10 times. However, as sequencing of genomes is very biased to certain parts of it and others are rarely sequenced, low coverage sequencing usually results in that only genes occurring in high copy numbers like the mitochondrial genes and nuclear ribosomal (18S and 28S) rRNA ones are sequenced well enough to be determined with certainty. In a way the high copy cream of the genome milk is skimmed off. Based on the mitochondrial and nuclear 18S and 28S genes, we could find that the molecular evidence supported the separation of the two subgenera and their elevation to genus status (Fig. 2). This was further support by morphological evidence, for example, in the male genitalia. Hence, in the paper we resurrected the genus status for Brachycampta and emended the status for Allodia.
How did I come involved in this paper? In my opinion, it shows the benefits one can get from evaluation procedures implemented in PhD project. At the NHM, the progress of a PhD project is annually evaluated by an internal committee, which does not comprise the supervising team. The goal is to provide an outside view on the project, have at least annual landmarks at which the PhD student and its supervising reflect upon the progress achieved and most importantly assist the PhD student in her/his goal to finish the PhD project successfully with by advice and help. During one of these meeting, we discussed a part of her project and I suggested that maybe a genome skimming approach would be efficient to accomplish the same goal. Afterwards Trude, Geir, Arild and I discussed it further. Trude did then a tremendous job pulling it off and I got partly involved in her nice project on Allodia.
Another more sad spin to the story is that the publication was substantially delayed due to bad editorial handling by Molecular Phylogenetics and Evolution. Before submission of her PHD thesis and her parental leave, Trude submitted the paper to MPE. Despite several inquiries about its progress, there it remained on the editor’s desk without any action (no editorial rejection or sending it out for review) until she came from her parental leave more than a year after submission. Hence, in the time, where the responsible MPE editor did nothing, Trude defended her thesis successfully, gave birth to a child and started settling into the new life as parent. At that time, we decided to withdraw the submission from MPE and send it to Systematic Entomology, where it eventually got published after a proper and thorough review process. With all due respect, to the burdens imposed on us due to the Covid pandemic, such long times without any decision are not acceptable. As we learned, we were not the only entomological paper affected by this. The editorial process at MPE as to improve to avoid such cases in the future.
Magnussen, T., Johnsen, A., Kjærandsen, J., Struck, T.H. and Søli, G.E.E. (2022), Molecular phylogeny of Allodia (Diptera: Mycetophilidae) constructed using genome skimming. Syst Entomol, 47: 267-281. https://doi.org/10.1111/syen.12529
![]()
